Chisio for BioPAX
Chisio is an open source generic graph editing toolkit
which was developed in Bilkent University. In this project we use Chisio for developing an open source framework for
visualizing, editing and analyzing BioPAX graphs.
Features:
- Displays BioPAX graphs as process diagrams.
- Uses compound graphs for showing complex molecules and cellular locations.
- Supports automated layout (Compound Spring Embedder).
- Maintains multiple views of the same BioPAX file.
- Can load and display many types of experimental data on the pathway diagram such as gene expression,
mutation, copy number variation, and mass spectrometry data.
- Facilitates querying Pathway Commons database for pathways and molecule neighborhoods.
- Supports merging two BioPAX files using merge facility of Paxtools.
- Discovers and displays possible causative paths of gene expressions based on the loaded expression data.
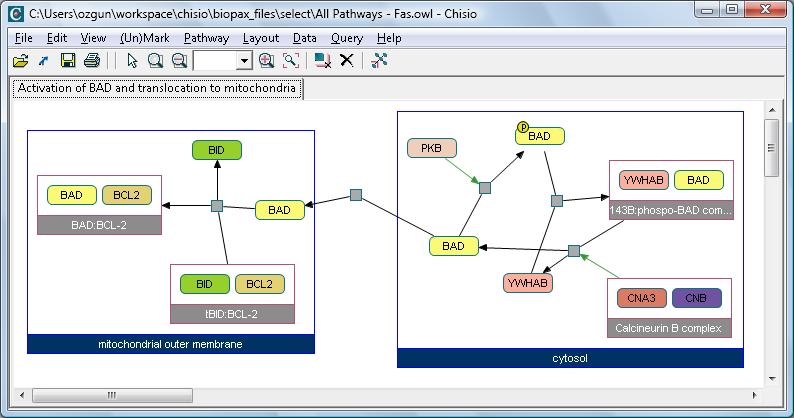
Below is a screenshot of the application displaying activation and translocation of BAD protein.
Chisio BioPAX Editor is still in alpha. The working copy of the source code can be retrieved using sourceforge CVS
service. Please follow these
guidelines for checking out the latest working copy.

